If you do not have access to one of our papers, contact us. We will be happy to send you a copy.
Virginia Tech
2025 Open Biol - A distinct phase of cyclin B (Cdc13) nuclear export at mitotic entry in S. pombe / Chethan SG, Rogers JM, Vijayakumari D, Williams W, Gligorovski V, Rahi SJ, Hauf S / DOI: 10.1098/rsob.250199
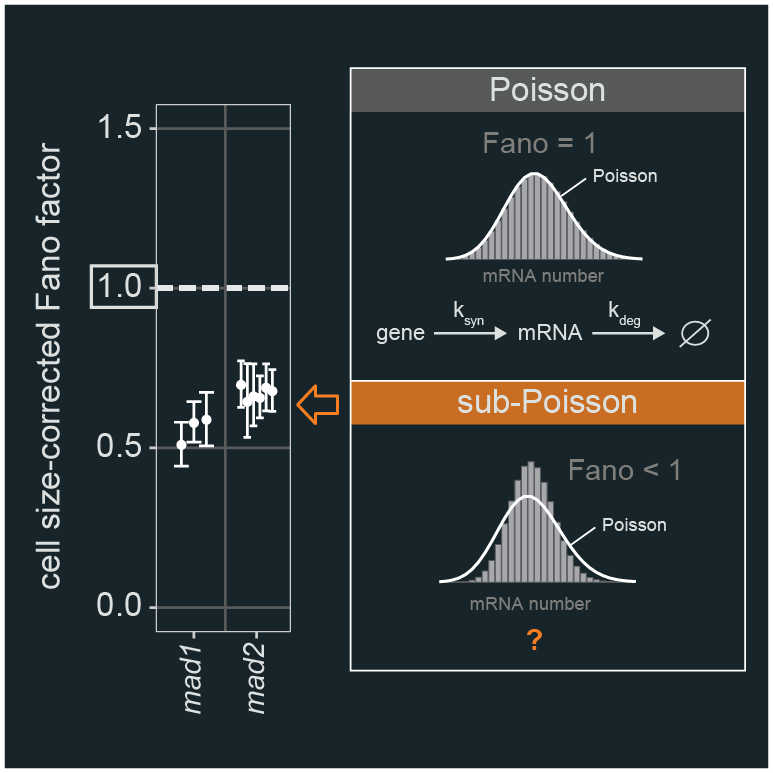
2023 Sci Adv - The minimal intrinsic stochasticity of constitutively expressed eukaryotic genes is sub-Poissonian / Weidemann DE, Holehouse J, Singh A, Grima R, Hauf S / DOI: 10.1126/sciadv.adh5138
2022 EMBO J - Mitotic checkpoint gene expression is tuned by codon usage bias / Esposito E, Weidemann DE, Rogers JM, Morton CM, Baybay EK, Chen J, Hauf S / DOI: 10.15252/embj.2021107896
2022 Cell Reports - Cdc48 influence on separase levels is independent of mitosis and suggests translational sensitivity of separase / Vijayakumari D, Müller J, Hauf S / DOI: 10.1016/j.celrep.2022.110554
2022 Nucleic Acids Res - Mutation and selection explain why many eukaryotic centromeric DNA sequences are often A + T rich / Barbosa AC, Xu Z, Karari K, Williams W, Hauf S, Brown WRA / DOI: 10.1093/nar/gkab1219
2020 Scientific Reports - Pomegranate: 2D segmentation and 3D reconstruction for fission yeast and other radially symmetric cells / Baybay EK, Esposito E, Hauf S / https://rdcu.be/b779h
2018 PLoS Comp Biol - Implications of alternative routes to APC/C inhibition by the mitotic checkpoint complex / Gross F, Bonaiuti P, Hauf S, Ciliberto A / DOI: 10.1371/journal.pcbi.1006449
2017 Cold Spring Harb Protoc - Construction, Growth, and Harvesting of Fission Yeast Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) Strains / Koch A, Bicho CC, Borek WE, Carpy A, Maček B, Hauf S, Sawin KE / DOI: 10.1101/pdb.prot091678
2017 Cold Spring Harb Protoc - Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) Technology in Fission Yeast / Maček B, Carpy A, Koch A, Bicho CC, Borek WE, Hauf S, Sawin KE / DOI: 10.1101/pdb.top079814
2017 Cold Spring Harb Protoc - Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC)-Based Quantitative Proteomics and Phosphoproteomics in Fission Yeast / Carpy A, Koch A, Bicho CC, Borek WE, Hauf S, Sawin KE, Maček B / DOI: 10.1101/pdb.prot091686
2017 Curr Biol - Different functionality of Cdc20 binding sites within the mitotic checkpoint complex / Sewart K, Hauf S / DOI: 10.1016/j.cub.2017.03.007
2017 eLife - Cell cycle: Micromanaging checkpoint proteins [Commentary] / Ciliberto A, Hauf S / DOI: 10.7554/eLife.25001
2016 Trends Cell Biol - Time to split up: Dynamics of chromosome separation / Kamenz J, Hauf S / DOI: 10.1016/j.tcb.2016.07.008
2016 Bioinformatics - MEMO - Multi-experiment mixture model analysis of censored data / Geissen EM, Hasenauer, J, Heinrich S, Hauf S, Theis FJ, Radde N / DOI: 10.1093/bioinformatics/btw190
2015 Mol Cell - Robust ordering of anaphase events by adaptive thresholds and competing degradation pathways / Kamenz J, Mihaljev T, Kubis A, Legewie S, Hauf S / DOI: 10.1016/j.molcel.2015.09.022
commentary by Malumbres, Dev Cell 2015
2014 SIViP - Graph-regularized 3D shape reconstruction from highly anisotropic and noisy images / Widmer C, Heinrich S, Drewe P, Lou X, Umrania S, Rätsch G/ DOI: 10.1007/s11760-014-0694-8
2014 Angew Chem Int Ed Engl - In vitro reconstitution of a cellular phase-transition process that involves the mRNA recapping machinery / Fromm SA, Kamenz J, Nöldeke ER, Neu A, Zocher G, Sprangers R / DOI: 10.1002/anie.201402885
2014 Mol Cell Proteomics - Absolute proteome and phosphoproteome dynamics during the cell cycle of fission yeast / Carpy A*, Krug K*, Graf S*, Koch A*, Popic S, Hauf S, Macek B / *equal contribution / DOI: 10.1074/mcp.M113.035824
2014 Curr Biol - Slow checkpoint activation kinetics as a safety device in anaphase / Kamenz J, Hauf S / DOI: 10.1016/j.cub.2014.02.005
commentary by Kops, Curr Biol 2014
article in BioTechniques
2014 EMBO rep - Mad1 contribution to checkpoint signalling goes beyond presenting Mad2 at kinetochores / Heinrich S, Sewart K, Windecker H, Langegger M, Schmidt N, Hustedt N, Hauf S / DOI: 10.1002/embr.201338114
commentary by Overlack, Krenn and Musacchio, EMBO rep 2014
Max Planck Campus, Tübingen
2013 Biochem Soc Trans - The spindle assembly checkpoint: progress and persistent puzzles / [mini-review] / Hauf S / DOI: 10.1042/BST20130240
2013 Nat Cell Biol - Determinants of robustness in spindle assembly checkpoint signalling / Heinrich S, Geissen EM, Kamenz J, Trautmann S, Widmer C, Drewe P, Knop M, Radde N, Hasenauer J, Hauf S / DOI: 10.1038/ncb2864
commentary by Subramanian and Kapoor, Nat Cell Biol 2013
featured on the cover
2012 J Cell Sci - Mph1 kinetochore localization is crucial and upstream in the hierarchy of spindle assembly checkpoint protein recruitment to kinetochores / Heinrich S, Windecker H, Hustedt N, Hauf S / DOI: 10.1242/jcs.110387
2012 ACS Chem Biol - A chemical genetic approach for covalent inhibition of analogue-sensitive aurora kinase / Koch A, Rode HB, Richters A, Rauh D, Hauf S / DOI: 10.1021/cb200465c
2012 EMBO J - The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex / Fromm SA, Truffault V, Kamenz J, Braun JE, Hoffmann NA, Izaurralde E, Sprangers R / DOI: 10.1038/emboj.2011.408
2011 Dev Cell - Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I / Sakuno T, Tanaka K, Hauf S, Watanabe Y / DOI: 10.1016/j.devcel.2011.08.012
2011 Sci Signal - Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast / Koch A, Krug K, Pengelley S, Macek B, Hauf S / DOI: 10.1126/scisignal.2001588
covered in Science Signaling podcast
2010 Eur J Cell Biol - Strategies for the identification of kinase substrates using analog-sensitive kinases / [review] / Koch A, Hauf S / DOI: 10.1016/j.ejcb.2009.11.024
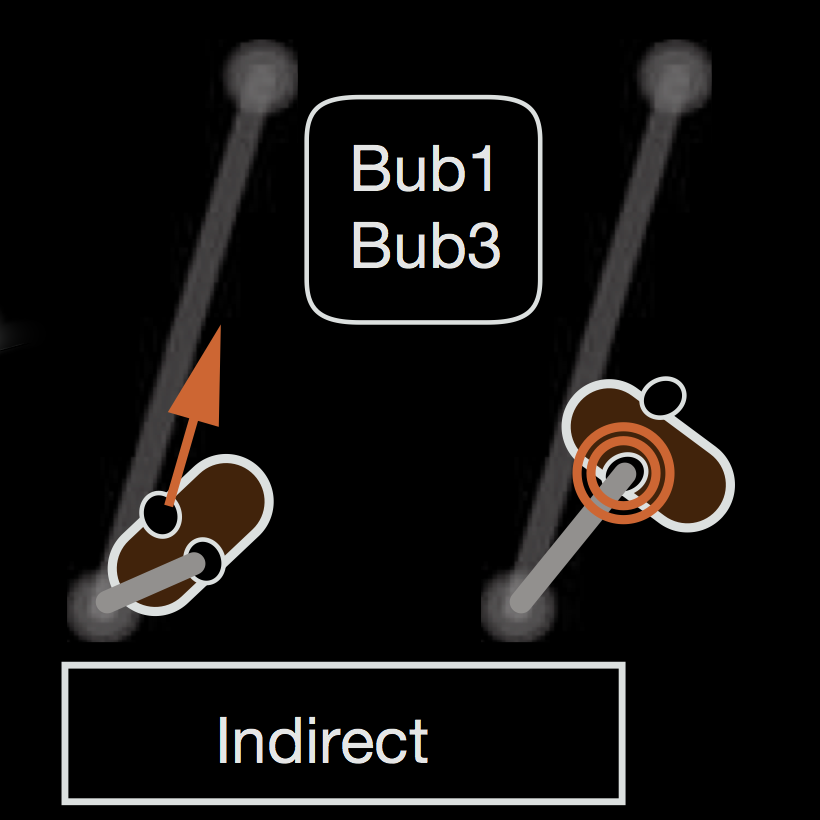
2009 EMBO rep - Bub1 and Bub3 promote the conversion from monopolar to bipolar chromosome attachment independently of shugoshin / Windecker H, Langegger M, Heinrich S, Hauf S / DOI: 10.1038/embor.2009.183
2008 Cell - Mps1 checks up on chromosome attachment / [commentary] / Hauf S / DOI: 10.1016/j.cell.2008.01.008
2007 EMBO J - Aurora controls sister kinetochore mono-orientation and homolog bi-orientation in meiosis-I / Hauf S*, Biswas A, Langegger M, Kawashima SA, Tsukahara T, Watanabe Y / *corresponding / DOI: 10.1038/sj.emboj.7601880
2007 Genes Dev - Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres / Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y / DOI: 10.1101/gad.1497307
Vienna / Tokyo
2005 PLoS Biol - Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2 / Hauf S*, Roitinger E*, Koch B*, Dittrich CM, Mechtler K, Peters JM / *equal contribution / DOI: 10.1371/journal.pbio.0030069
2005 Curr Biol - Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization / Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y / DOI: 10.1016/j.cub.2004.12.044
2005 Cell - Kinetochore orientation in mitosis and meiosis / [review] / Hauf S, Watanabe Y / DOI: 10.1016/j.cell.2004.10.014
2004 Curr Biol - Regulation of sister chromatid cohesion between chromosome arms / Giménez-Abián JF, Sumara I, Hirota T, Hauf S, Gerlich D, de la Torre C, Ellenberg J, Peters JM / DOI: 10.1016/j.cub.2004.06.052
2003 Cell Cycle - Fine tuning of kinetochore function by phosphorylation / [commentary] / Hauf S / DOI: 10.4161/cc.2.3.367
2003 J Cell Biol - The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint / Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM / DOI: 10.1083/jcb.200208092
2001 Science - Cohesin cleavage by separase required for anaphase and cytokinesis in human cells / Hauf S, Waizenegger IC, Peters JM / DOI: 10.1126/science.1061376
2000 Cell - Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase / Waizenegger IC, Hauf S, Meinke A, Peters JM / DOI: 10.1016/S0092-8674(00)00132-X